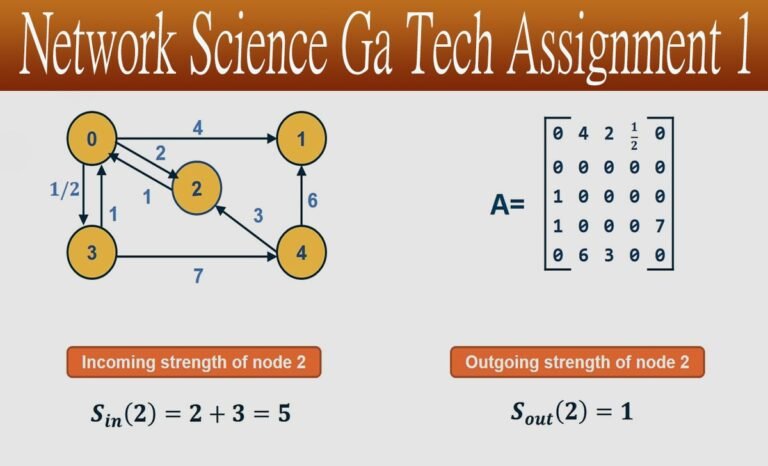
Introduction
If you are tackling network science ga tech assignment 1, you are in a great place. This guide walks you through key ideas and practical steps. I use plain words and short sentences. Each section explains one helpful topic. You will see how to read the task, set up tools, analyze graphs, and present results. I include tips on NetworkX, Python, and visualization. I also add writing and time management tips for students. The goal is to make the assignment feel simple and doable. Follow the steps, test early, and document everything. If you are stuck, the structure below will show where to check next. Use this guide as a map for a clear, reproducible solution to network science ga tech assignment 1.
Contents
Read the assignment carefully and parse the tasks
Start by reading network science ga tech assignment 1 with a highlighter. Note required outputs and formats. List the tasks in plain words. Mark due dates and any sample files. Break the assignment into steps. For example, steps may include loading data, computing degree distributions, and plotting a network. Check for required libraries like NetworkX or numpy. Note if they ask for code, figures, and a short report. If the assignment has rubric points, match each deliverable to the rubric. This early mapping saves time. It helps you avoid missing parts. Keep a checklist and tick each task as you finish it. That makes your work organized and stress-free.
Set up a clean environment and required tools
Before you code, make a clean project folder. Install Python and virtual environment tools. Use venv or conda to isolate packages. Install NetworkX, pandas, matplotlib, and seaborn. If the course uses Jupyter, create a notebook for exploration. Save raw data in a data/ folder and scripts in src/. Track versions in a requirements.txt file. This setup helps reproducibility. If you share code, others can re-run it easily. For network science ga tech assignment 1, using a consistent environment avoids “it works on my machine” problems. Back up your work often and commit changes if you use git. A tidy environment makes later analysis smoother.
Understand the data: nodes, edges, and formats
The core of network science ga tech assignment 1 is the graph data. Check whether files are edge lists, adjacency matrices, or JSON. Read the sample data first. Note node labels, weighted edges, and timestamps if present. If edges have types or directions, record that detail. Use pandas to preview CSV files. Confirm whether self-loops or parallel edges exist. Decide if you should simplify the graph by removing duplicates or isolating the giant component. Document any cleaning steps. Clear data understanding prevents wrong analyses. A small note in your report about data quirks shows care and builds trust.
Build the graph in Python using NetworkX
NetworkX is a friendly library for network tasks. Start by importing NetworkX and reading your file. Use nx.read_edgelist or nx.from_pandas_edgelist as needed. If edges are weighted, include the weight field. For directed graphs, use nx.DiGraph(). Add node attributes if helpful, like categories or dates. After building the graph, print basic stats: number of nodes, number of edges, and graph density. Visualize a small sample to check structure. Save the script as build_graph.py and comment each step. For network science ga tech assignment 1, clear, reproducible graph construction is key to valid results.
Compute degree and degree distribution correctly
Degree gives basic insight into node connectivity. Compute degree for each node with G.degree(). For directed graphs, use in_degree() and out_degree(). Plot the degree distribution as a histogram with log bins if needed. Fit a power-law or log-normal curve cautiously and report fit quality. Compute mean, median, variance, and identify hubs by sorting degrees. Use cumulative distribution plots where helpful. For network science ga tech assignment 1, show both numeric summary and clear plot images. Explain what the degree distribution says about network heterogeneity and role of hub nodes.
Centrality measures: what to compute and why
Centrality helps identify important nodes. For network science ga tech assignment 1, compute at least degree, betweenness, closeness, and eigenvector centrality. Use NetworkX functions like nx.betweenness_centrality(). Normalize values where standard. Interpret each measure: degree shows local connections, betweenness shows brokering roles, closeness shows accessibility, and eigenvector shows influence within dense hubs. Rank nodes for each metric and look for agreement among measures. If disagreements appear, explain why. Present tables of top nodes and short comments on their roles. Do not compute unnecessary measures without interpretation.
Clustering coefficient and local structure
Clustering coefficient measures triangle density around nodes. Compute local clustering with nx.clustering() and the average clustering coefficient with nx.average_clustering(). For directed or weighted graphs, use the correct variants. Explain what high clustering means: local cohesion and redundancy. Compare clustering to random graph baselines. You can generate Erdős–Rényi graphs with the same number of nodes and edges and compare average clustering. A contrast like this highlights structure beyond randomness. For network science ga tech assignment 1, include such comparisons to strengthen conclusions.
Community detection and modularity insights
Community detection groups nodes into modules. Use algorithms like Louvain, Girvan-Newman, or spectral clustering. NetworkX may need external libraries like python-louvain for Louvain. Compute modularity to assess the partition quality. Visualize communities with node colors and a legend. Report sizes and central nodes in each community. Discuss whether communities correspond to known node attributes, like categories or geography, when available. For network science ga tech assignment 1, pick one or two algorithms and justify your choice. Show code and results that are easy to reproduce.
Path lengths, diameter, and small-world properties
Compute shortest-path lengths with nx.shortest_path_length() and find the average shortest path. Find the diameter of the largest connected component with nx.diameter(). Compare the average path length and clustering to random graphs to assess small-world behavior. A small-world network typically has high clustering and low average path length. Present a concise table comparing metrics for your real graph and random baselines. Explain implications: short paths indicate fast spread or short distances between nodes. For network science ga tech assignment 1, include interpretation and short code snippets to show the exact method.
PageRank and flow-based importance
PageRank measures node importance via random-walk ideas. Use nx.pagerank() with a damping factor like 0.85. If edges are weighted, pass the weight attribute. Rank nodes by PageRank and compare to degree or eigenvector centrality. Discuss why PageRank might highlight different nodes. For example, nodes with fewer but well-connected neighbors can rank higher. Include a short visualization showing PageRank values as node sizes. For network science ga tech assignment 1, such contrasts enrich your narrative and show deeper understanding of flow-based metrics.
Visualizing the network effectively
Good visuals make analysis clear. Use matplotlib with NetworkX or export to Gephi for interactive layout work. Choose layouts like spring (force-directed) for general structure or circular for small graphs. Color nodes by community or centrality and size them by degree or PageRank. Keep labels minimal to avoid clutter. For larger graphs, show subgraph zoom-ins for clarity. Save high-resolution PNGs for the report. For network science ga tech assignment 1, include at least two visualizations: a full-network overview and a zoomed community or hub view. Caption each image with a short explanation.
Statistical testing and null models
To show that your findings are significant, use null models. Generate random graphs with the same degree sequence via the configuration model. Compare metrics like clustering, modularity, and path length across many random instances. Compute p-values for observed metrics versus the random ensemble. This testing demonstrates whether observed structure is likely by chance. For network science ga tech assignment 1, a simple set of 100 random graphs usually suffices to show trends. Report the mean and standard deviation for null metrics to support claims.
Reproducible reporting and code sharing
Your submission should include code and a short report. Put code in a notebooks/ or scripts/ folder. Add a README with execution steps and requirements. Use clear function names and comments. Include figures with captions and a concise results section. Zip the project or push to a GitHub repo and include a link. For network science ga tech assignment 1, reproducibility is often graded. Make it easy for graders to run your notebook. Document random seeds and versions. A reproducible repo increases credibility and earns trust.
Common pitfalls and how to avoid them
Many students rush graphs without cleaning data. Missing node attributes or duplicates give wrong answers. Another pitfall is overfitting fancy metrics without interpretation. Always explain why a metric matters. Avoid plotting unreadable graphs with thousands of labels. Use baselines and null models to check significance. Also watch performance: very large graphs need sparse algorithms and efficient memory handling. For network science ga tech assignment 1, test code on small samples before scaling. Keep outputs concise and focused on rubric items.
Time management and how to split the work
Break the assignment into timed blocks. Day 1: read and set up environment. Day 2: data cleaning and graph building. Day 3: compute core metrics and plots. Day 4: run null models and refine figures. Day 5: write the report and bundle code. Use shorter blocks for testing code and debugging. Keep a checklist and mark items complete. Ask teaching staff early if you hit blockers. For network science ga tech assignment 1, starting early gives you time to test assumptions and add polish, which often raises grades.
Tips for writing the final report and presentation
Write a concise abstract that answers the main questions. Include a methods section with software and versions. Present results with clear figures and short interpretation paragraphs. Add a limitations section and suggestions for future work. Keep language simple and direct. Use bullet lists for key takeaways. For presentations, prepare 5–7 slides that summarize the problem, methods, main results, and implications. Rehearse aloud and time your talk. For network science ga tech assignment 1, clear explanation beats fancy jargon.
Extra credit ideas and extensions
If you have time, add dynamic analysis like temporal graphs or epidemic simulations. Try multilayer networks if data supports it. Implement community detection comparisons or sentiment-based node attributes. Use Gephi or D3.js for interactive visuals. Another extension is to apply machine learning to predict missing links. These extras show initiative and deeper curiosity. For network science ga tech assignment 1, pick one extension that fits the dataset and finish it well, rather than trying many partial extras.
Conclusion and final checklist
To finish network science ga tech assignment 1, follow this quick checklist: parse tasks, set up environment, clean data, build the graph, compute core metrics, run null models, make clear plots, and write a concise report. Zip your code, add a README, and include a reproducible notebook. Double-check file formats and naming conventions. Run through the rubric to ensure you covered required items. Submit early if possible to catch upload problems. A careful, documented approach often yields better results than last-minute complexity. Good luck, and enjoy exploring the network!
FAQs — common questions students ask about assignment 1
FAQ 1 — What are the essential libraries to install?
Install Python 3.8+ and create a virtual environment. Add NetworkX, pandas, numpy, matplotlib, seaborn, and scipy. For community detection, install python-louvain or community package. For large graphs, consider igraph or graph-tool if allowed. Keep a requirements.txt file. This list covers most tasks in network science ga tech assignment 1 and helps reproducible runs.
FAQ 2 — How do I handle a graph with thousands of nodes?
For large graphs, use sparse data structures and avoid heavy visualizations. Compute metrics incrementally and use sampling for plots. Use networkx carefully; if it slows, try igraph or graph-tool. For path computations, restrict to the giant component or sample node pairs. Keep memory usage in mind and test on smaller subsets first. These steps let you complete network science ga tech assignment 1 on realistic hardware.
FAQ 3 — Should I include raw data files in my submission?
Include raw data if permitted and file sizes are reasonable. Otherwise provide a link to the data source with access instructions. Always include processing scripts that transform raw data into the graph. For network science ga tech assignment 1, show enough that graders can reproduce your steps without hunting for missing files.
FAQ 4 — How do I pick which centrality to trust?
No single centrality fits every purpose. Use degree for local influence, betweenness for bridges, closeness for access, and eigenvector/PageRank for global influence. Compare top nodes across metrics and explain differences. The choice depends on the question you ask. For network science ga tech assignment 1, justify your selections in the report.
FAQ 5 — What if my plots look messy or unreadable?
Reduce node labels and simplify colors. Show subgraphs for clarity. Use node aggregation or community summaries. Export high-resolution PNGs or PDFs. For interactive display, use Gephi or web tools. Clear captions help graders interpret visuals. Clean, focused figures improve understanding for network science ga tech assignment 1.
FAQ 6 — How long should the report be?
Follow any page limits in the rubric. A concise report often runs 4–8 pages with figures. Include an abstract, methods, results, and short discussion. Append code or link to a GitHub repo. Clarity and reproducibility matter more than length. A focused and well-documented report helps graders see your work on network science ga tech assignment 1.Introduction
If you are tackling network science ga tech assignment 1, you are in a great place. This guide walks you through key ideas and practical steps. I use plain words and short sentences. Each section explains one helpful topic. You will see how to read the task, set up tools, analyze graphs, and present results. I include tips on NetworkX, Python, and visualization. I also add writing and time management tips for students. The goal is to make the assignment feel simple and doable. Follow the steps, test early, and document everything. If you are stuck, the structure below will show where to check next. Use this guide as a map for a clear, reproducible solution to network science ga tech assignment 1.
Read the assignment carefully and parse the tasks
Start by reading network science ga tech assignment 1 with a highlighter. Note required outputs and formats. List the tasks in plain words. Mark due dates and any sample files. Break the assignment into steps. For example, steps may include loading data, computing degree distributions, and plotting a network. Check for required libraries like NetworkX or numpy. Note if they ask for code, figures, and a short report. If the assignment has rubric points, match each deliverable to the rubric. This early mapping saves time. It helps you avoid missing parts. Keep a checklist and tick each task as you finish it. That makes your work organized and stress-free.
Set up a clean environment and required tools
Before you code, make a clean project folder. Install Python and virtual environment tools. Use venv or conda to isolate packages. Install NetworkX, pandas, matplotlib, and seaborn. If the course uses Jupyter, create a notebook for exploration. Save raw data in a data/ folder and scripts in src/. Track versions in a requirements.txt file. This setup helps reproducibility. If you share code, others can re-run it easily. For network science ga tech assignment 1, using a consistent environment avoids “it works on my machine” problems. Back up your work often and commit changes if you use git. A tidy environment makes later analysis smoother.
Understand the data: nodes, edges, and formats
The core of network science ga tech assignment 1 is the graph data. Check whether files are edge lists, adjacency matrices, or JSON. Read the sample data first. Note node labels, weighted edges, and timestamps if present. If edges have types or directions, record that detail. Use pandas to preview CSV files. Confirm whether self-loops or parallel edges exist. Decide if you should simplify the graph by removing duplicates or isolating the giant component. Document any cleaning steps. Clear data understanding prevents wrong analyses. A small note in your report about data quirks shows care and builds trust.
Build the graph in Python using NetworkX
NetworkX is a friendly library for network tasks. Start by importing NetworkX and reading your file. Use nx.read_edgelist or nx.from_pandas_edgelist as needed. If edges are weighted, include the weight field. For directed graphs, use nx.DiGraph(). Add node attributes if helpful, like categories or dates. After building the graph, print basic stats: number of nodes, number of edges, and graph density. Visualize a small sample to check structure. Save the script as build_graph.py and comment each step. For network science ga tech assignment 1, clear, reproducible graph construction is key to valid results.
Compute degree and degree distribution correctly
Degree gives basic insight into node connectivity. Compute degree for each node with G.degree(). For directed graphs, use in_degree() and out_degree(). Plot the degree distribution as a histogram with log bins if needed. Fit a power-law or log-normal curve cautiously and report fit quality. Compute mean, median, variance, and identify hubs by sorting degrees. Use cumulative distribution plots where helpful. For network science ga tech assignment 1, show both numeric summary and clear plot images. Explain what the degree distribution says about network heterogeneity and role of hub nodes.
Centrality measures: what to compute and why
Centrality helps identify important nodes. For network science ga tech assignment 1, compute at least degree, betweenness, closeness, and eigenvector centrality. Use NetworkX functions like nx.betweenness_centrality(). Normalize values where standard. Interpret each measure: degree shows local connections, betweenness shows brokering roles, closeness shows accessibility, and eigenvector shows influence within dense hubs. Rank nodes for each metric and look for agreement among measures. If disagreements appear, explain why. Present tables of top nodes and short comments on their roles. Do not compute unnecessary measures without interpretation.
Clustering coefficient and local structure
Clustering coefficient measures triangle density around nodes. Compute local clustering with and the average clustering coefficient with For directed or weighted graphs, use the correct variants. Explain what high clustering means: local cohesion and redundancy. Compare clustering to random graph baselines. You can generate Erdős–Rényi graphs with the same number of nodes and edges and compare average clustering. A contrast like this highlights structure beyond randomness. For network science ga tech assignment 1, include such comparisons to strengthen conclusions.
Community detection and modularity insights
Community detection groups nodes into modules. Use algorithms like Louvain, Girvan-Newman, or spectral clustering. NetworkX may need external libraries like python-louvain for Louvain. Compute modularity to assess the partition quality. Visualize communities with node colors and a legend. Report sizes and central nodes in each community. Discuss whether communities correspond to known node attributes, like categories or geography, when available. For network science ga tech assignment 1, pick one or two algorithms and justify your choice. Show code and results that are easy to reproduce.
Path lengths, diameter, and small-world properties
Compute shortest-path lengths with and find the average shortest path. Find the diameter of the largest connected component with . Compare the average path length and clustering to random graphs to assess small-world behavior. A small-world network typically has high clustering and low average path length. Present a concise table comparing metrics for your real graph and random baselines. Explain implications: short paths indicate fast spread or short distances between nodes. For network science ga tech assignment 1, include interpretation and short code snippets to show the exact method.
PageRank and flow-based importance
PageRank measures node importance via random-walk ideas. Use with a damping factor like 0.85. If edges are weighted, pass the weight attribute. Rank nodes by PageRank and compare to degree or eigenvector centrality. Discuss why PageRank might highlight different nodes. For example, nodes with fewer but well-connected neighbors can rank higher. Include a short visualization showing PageRank values as node sizes. For network science ga tech assignment 1, such contrasts enrich your narrative and show deeper understanding of flow-based metrics.
Visualizing the network effectively
Good visuals make analysis clear. Use with NetworkX or export to Gephi for interactive layout work. Choose layouts like spring (force-directed) for general structure or circular for small graphs. Color nodes by community or centrality and size them by degree or PageRank. Keep labels minimal to avoid clutter. For larger graphs, show subgraph zoom-ins for clarity. Save high-resolution PNGs for the report. For network science ga tech assignment 1, include at least two visualizations: a full-network overview and a zoomed community or hub view. Caption each image with a short explanation.
Statistical testing and null models
To show that your findings are significant, use null models. Generate random graphs with the same degree sequence via the configuration model. Compare metrics like clustering, modularity, and path length across many random instances. Compute p-values for observed metrics versus the random ensemble. This testing demonstrates whether observed structure is likely by chance. For network science ga tech assignment 1, a simple set of 100 random graphs usually suffices to show trends. Report the mean and standard deviation for null metrics to support claims.
Reproducible reporting and code sharing
Your submission should include code and a short report. Put code in a folder. Add a README with execution steps and requirements. Use clear function names and comments. Include figures with captions and a concise results section. Zip the project or push to a GitHub repo and include a link. For network science ga tech assignment 1, reproducibility is often graded. Make it easy for graders to run your notebook. Document random seeds and versions. A reproducible repo increases credibility and earns trust.
Common pitfalls and how to avoid them
Many students rush graphs without cleaning data. Missing node attributes or duplicates give wrong answers. Another pitfall is overfitting fancy metrics without interpretation. Always explain why a metric matters. Avoid plotting unreadable graphs with thousands of labels. Use baselines and null models to check significance. Also watch performance: very large graphs need sparse algorithms and efficient memory handling. For network science ga tech assignment 1, test code on small samples before scaling. Keep outputs concise and focused on rubric items.
Time management and how to split the work
Break the assignment into timed blocks. Day 1: read and set up environment. Day 2: data cleaning and graph building. Day 3: compute core metrics and plots. Day 4: run null models and refine figures. Day 5: write the report and bundle code. Use shorter blocks for testing code and debugging. Keep a checklist and mark items complete. Ask teaching staff early if you hit blockers. For network science ga tech assignment 1, starting early gives you time to test assumptions and add polish, which often raises grades.
Tips for writing the final report and presentation
Write a concise abstract that answers the main questions. Include a methods section with software and versions. Present results with clear figures and short interpretation paragraphs. Add a limitations section and suggestions for future work. Keep language simple and direct. Use bullet lists for key takeaways. For presentations, prepare 5–7 slides that summarize the problem, methods, main results, and implications. Rehearse aloud and time your talk. For network science ga tech assignment 1, clear explanation beats fancy jargon.
Extra credit ideas and extensions
If you have time, add dynamic analysis like temporal graphs or epidemic simulations. Try multilayer networks if data supports it. Implement community detection comparisons or sentiment-based node attributes. Use Gephi or D3.js for interactive visuals. Another extension is to apply machine learning to predict missing links. These extras show initiative and deeper curiosity. For network science ga tech assignment 1, pick one extension that fits the dataset and finish it well, rather than trying many partial extras.
Conclusion
To finish network science ga tech assignment 1, follow this quick checklist: parse tasks, set up environment, clean data, build the graph, compute core metrics, run null models, make clear plots, and write a concise report. Zip your code, add a README, and include a reproducible notebook. Double-check file formats and naming conventions. Run through the rubric to ensure you covered required items. Submit early if possible to catch upload problems. A careful, documented approach often yields better results than last-minute complexity. Good luck, and enjoy exploring the network!
FAQs
FAQ 1 — What are the essential libraries to install?
Install Python 3.8+ and create a virtual environment. Add NetworkX, pandas, numpy, matplotlib, seaborn, and scipy. For community detection, install python-louvain or community package. For large graphs, consider igraph or graph-tool if allowed. Keep a requirements.txt file. This list covers most tasks in network science ga tech assignment 1 and helps reproducible runs.
FAQ 2 — How do I handle a graph with thousands of nodes?
For large graphs, use sparse data structures and avoid heavy visualizations. Compute metrics incrementally and use sampling for plots. Use networkx carefully; if it slows, try igraph or graph-tool. For path computations, restrict to the giant component or sample node pairs. Keep memory usage in mind and test on smaller subsets first. These steps let you complete network science ga tech assignment 1 on realistic hardware.
FAQ 3 — Should I include raw data files in my submission?
Include raw data if permitted and file sizes are reasonable. Otherwise provide a link to the data source with access instructions. Always include processing scripts that transform raw data into the graph. For network science ga tech assignment 1, show enough that graders can reproduce your steps without hunting for missing files.
FAQ 4 — How do I pick which centrality to trust?
No single centrality fits every purpose. Use degree for local influence, betweenness for bridges, closeness for access, and eigenvector/PageRank for global influence. Compare top nodes across metrics and explain differences. The choice depends on the question you ask. For network science ga tech assignment 1, justify your selections in the report.
FAQ 5 — What if my plots look messy or unreadable?
Reduce node labels and simplify colors. Show subgraphs for clarity. Use node aggregation or community summaries. Export high-resolution PNGs or PDFs. For interactive display, use Gephi or web tools. Clear captions help graders interpret visuals. Clean, focused figures improve understanding for network science ga tech assignment 1.
FAQ 6 — How long should the report be?
Follow any page limits in the rubric. A concise report often runs 4–8 pages with figures. Include an abstract, methods, results, and short discussion. Append code or link to a GitHub repo. Clarity and reproducibility matter more than length. A focused and well-documented report helps graders see your work on network science ga tech assignment 1.